title: “PREDICTING RISK FACTORS OF OSTEOPOROTIC FRACTURES IN UNITED STATES ADULTS” output: github_document —
```{r setup, include=FALSE} knitr::opts_chunk$set(echo = TRUE)
## INTRODUCTION
Osteoporosis is a common chronic disease of public health concern that results in fragility fractures affecting one in three women and one in five men over age 50 years. (IOF, 2020) This imposes a burden on the individuals and their caretakers result in morbidity and reduced quality of life. Individuals who sustained osteoporotic fractures endure back pain, loss of height, loss of mobility and many other adverse effects on the quality of their lives. Over 1.5 million fractures per year occurs due to osteoporosis with 250,000 hip fracture, 500,000 hospitalizations, 800,000 emergency room visits, 2,600,000 physician visits, and 180,000 nursing home placements. This cost the Unites states $20 billion in 2015 (Rodan, 2000). Hip fractures are a leading cause of morbidity and loss of independence in the older population and a 1-year mortality of 20% (Johnell,2006). Osteoporotic fractures had resulted in an estimated loss of 5.8 million disability adjusted life years (DALYs) yearly (Johnell,2006).
These staggering cost on the health care system and the excruciating burden on the patient and the healthcare system needs to be examined by public health professionals and medical personnel. Osteoporosis can be treated, and fractures prevented.
Osteoporosis is a silent disease that results from imbalance between bone resorption and bone formation.
This is a public health emergency in the aging population that requires an urgent attention particularly in the light of the COVID- 19 pandemic where certain part of the health sector is neglected to attend to the more acute conditions. Early diagnosis of osteoporosis prior to fractures and prompt initiation of treatment and intervention methods are useful to reduce fragility fracture incidences through accurate prediction of fracture risks.
## DATA DESCRIPTION
Data source is the National Health and Nutrition Examination Survey (NHANES) 2017- 2018, a nationally representative, cross-sectional survey designed to assess the health and nutritional status of adults and children across the United States. NHANES is a major program of the National Center for Health Statistics (NCHS) and is responsible for producing vital and health statistics for the Nation. (NCHS, 2017). The survey examines a national representative sample of individuals located in counties across the country.
This study uses data from the National Health and Nutrition Examination Survey, 2017-2018 (NHANES 2017-2018), collected between January 2017 and December 2018.
#SKILLS- knitr, plyr, dplyr, VIM, mice, ggplot2, caret, glm, cross validation, logistics regression, decision tree, rendom forest, Lasso/ Ridge regression, Neural Network,SVM, Bagging, Boosting
#install.packages(“nhanesA”) library(nhanesA) require(knitr) #for nice tables #install.packages(“SASxport”) require(SASxport)
#nhanesTables(data_group= “DEMO”, year=2017)
#kable(nhanesTables(data_group= “LAB”, year=2017)) #nicer table #kable(nhanesTables(data_group= “DEMO”, year=2017)) #kable(nhanesTables(data_group= “DIET”, year=2017)) #kable(nhanesTables(data_group= “Q”, year=2017)) #kable(nhanesTables(data_group= “EXAM”, year=2017))
#kable(nhanesTableVars(data_group=’DEMO’, nh_table= ‘DEMO_J’, namesonly=FALSE))
DATA WRANGLING
#selecting variables demo <- nhanes(‘DEMO_J’) demo1 <- demo[c(“SEQN”, “RIAGENDR”, “RIDAGEYR”, “RIDRETH3”, “DMDEDUC2”, “DMDMARTL”, “INDFMPIR”)] demo_vars <-names(demo1) demo2 <- nhanesTranslate (“DEMO_J”, demo_vars, data=demo1)
diet<- nhanes(‘DSQTOT_J’) diet1 <- diet[c(“SEQN”,”DSQTVD”)] diet_vars <-names(diet1) diet2 <- nhanesTranslate (“DSQTOT_J”, diet_vars, data=diet1) head(diet2)
#”DR1TPROT”- protein intake
smok<- nhanes(‘SMQ_J’) smok1 <- smok[c(“SEQN”, “SMQ040”, “SMQ020”)] smok_vars <-names(smok1) smok2 <- nhanesTranslate (“SMQ_J”, smok_vars, data=smok1) head(smok2)
alc<- nhanes(‘ALQ_J’) alc1 <- alc[c(“SEQN”, “ALQ130”,”ALQ111”)] alc_vars <-names(alc1) alc2 <- nhanesTranslate (“ALQ_J”, alc_vars, data=alc1) head(alc2)
diab<- nhanes(‘DIQ_J’) diab1 <- diab[c(“SEQN”, “DIQ010”)] diab_vars <-names(diab1) diab2 <- nhanesTranslate (“DIQ_J”, diab_vars, data=diab1) head(diab2)
mcq<- nhanes(‘MCQ_J’) mcq1 <- mcq [c(“SEQN”, “MCQ160A”,”MCQ160F”)] mcq_vars <-names(mcq1) mcq2 <- nhanesTranslate (“MCQ_J”, mcq_vars, data=mcq1) head(mcq2)
osteo<- nhanes(‘OSQ_J’) osteo1<- osteo [c( “SEQN”, “OSQ010A”,”OSQ010B”, “OSQ010C”,”OSQ060”,”OSQ130”, “OSQ150”)] osteo_vars <-names(osteo1) osteo2 <- nhanesTranslate (“OSQ_J”, osteo_vars, data=osteo1) head(osteo2)
preg<- nhanes(‘RHQ_J’) preg1<- preg [c( “SEQN”, “RHQ540”)] preg_vars <-names(preg1) preg2 <- nhanesTranslate (“RHQ_J”, preg_vars, data=preg1) head(preg2)
bmi<- nhanes(‘WHQ_J’) bmi1<- bmi [c( “SEQN”, “WHD020”,”WHD010”)] bmi_vars <-names(bmi1) bmi2 <- nhanesTranslate (“WHQ_J”, bmi_vars, data=bmi1) head(bmi2)
fem<- nhanes(‘DXXFEM_J’) fem1<- fem [c( “SEQN”, “DXXNKBMD”)] fem_vars <-names(fem1) fem2 <- nhanesTranslate (“DXXFEM_J”, fem_vars, data=fem1) head(fem2)
spine<- nhanes(‘DXXSPN_J’) spine1<- spine [c( “SEQN”, “DXXOSBMD”)] spine_vars <-names(spine1) spine2 <- nhanesTranslate (“DXXSPN_J”, spine_vars, data=spine1) head(spine2)
bio<- nhanes(‘BIOPRO_J’) bio1<- bio [c( “SEQN”, “LBXSCA”)] bio_vars <-names(bio1) bio2 <- nhanesTranslate (“BIOPRO_J”, bio_vars, data=bio1) head(bio2)
pa<- nhanes(‘PAQ_J’) pa1<- pa [c(“SEQN”, “PAQ605”, “PAQ620”, “PAQ635”, “PAQ650”, “PAQ665”, “PAD680”)] pa_vars <-names(pa1) pa2 <- nhanesTranslate (“PAQ_J”, pa_vars, data=pa1) head(pa2)
#merge
#data<- merge(demo2, diet2, by= c(“SEQN”), all=TRUE) #data1<- merge(data, smok2, by= c(“SEQN”), all=TRUE) #data2<- merge(data1, alc2, by= c(“SEQN”), all=TRUE) #data3<- merge(data2, diab2, by= c(“SEQN”), all=TRUE) #data4<- merge(data3, mcq2, by= c(“SEQN”), all=TRUE) #data5<- merge(data4, osteo2, by= c(“SEQN”), all=TRUE) #data6<- merge(data5, preg2, by= c(“SEQN”), all=TRUE) #data7<- merge(data6, bmi2, by= c(“SEQN”), all=TRUE) #data8<- merge(data7, fem2, by= c(“SEQN”), all=TRUE) #data9<- merge(data8, spine2, by= c(“SEQN”), all=TRUE) #data10<- merge(data9, tbmd2, by= c(“SEQN”), all=TRUE) #dim(data10) #head(data10)
Merging all at once
require(plyr) require(dplyr) data<-join_all(list(demo2, diet2, smok2, alc2, diab2, mcq2, osteo2, preg2, bmi2, fem2, spine2, bio2, pa2), by=”SEQN”, type=”full”) dim(data) # 9254 33
#saving data for late use getwd() save(data, file=”Project21.RData”)
#loading saved data load(“Project21.RData”) names(data) summary(data)
#subseting the data dataa<- subset(data,data$RIDAGEYR >=50, na.rm=TRUE) #restricting to age>=50, removing missing value dim(dataa) #3069 33 str(dataa) summary(dataa)
############################################################# #Femoral neck bmd #DXXNKBMD dat<- subset(dataa,dataa$DXXNKBMD >=0.1440, na.rm=TRUE) # restricting to those without missing values removing missing value for femoral BMD. summary(dat$DXXNKBMD) dim(dat)
#Fractures #Broken or fractured HIP,WRIST SPINE #renaming variables dat$Hipfrac [dat$OSQ010A==’No’]<-0 dat$Hipfrac [dat$OSQ010A==”Don’t know”]<-0 dat$Hipfrac [dat$OSQ010A==’Missing’]<-0 dat$Wristfrac [dat$OSQ010B==’No’]<-0 dat$Wristfrac [dat$OSQ010B==”Don’t know”]<-0 dat$Wristfrac [dat$OSQ010B==’Missing’]<-0 dat$spinefrac [dat$OSQ010C==’No’]<-0 dat$spinefrac [dat$OSQ010C==”Don’t know”]<-0 dat$spinefrac [dat$OSQ010C==’Missing’]<-0 dat$Hipfrac [dat$OSQ010A==’Yes’]<-1 dat$Wristfrac [dat$OSQ010B==’Yes’]<-1 dat$spinefrac [dat$OSQ010C==’Yes’]<-1
dat$frac [dat$Hipfrac==1]<-“Yes” dat$frac [dat$Hipfrac==0]<-“No” dat$frac [dat$Wristfrac==1]<-“Yes” dat$frac [dat$Wristfrac==0]<-“No” dat$frac [dat$spinefrac==1]<-“Yes” dat$frac [dat$spinefrac==0]<-“No”
table(dat$Hipfrac, useNA = “always”) table(dat$Wristfrac, useNA = “always”) table(dat$spinefrac, useNA = “always”)
table(dat$frac, useNA = “always”) frac <- na.omit(dat$frac)
#binary variable
#Gender #”RIAGENDR” - Gender
names(dat)
dat$Gender [dat$RIAGENDR==’Male’]<-‘Male’ dat$Gender [dat$RIAGENDR==’Female’]<-‘Female’ dat$Gender [dat$RIAGENDR==’Missing’]<-‘NA’
table(dat$Gender, useNA = “always”)
pie(table(dat$Gender), clockwise = TRUE, main=”Gender Distribution”, radius = 1,col=rainbow(2))
##Age #RIDAGEYR-
dat$age<-cut(dat$RIDAGEYR, c(-Inf,60,70, Inf),labels=c(‘50-59’,’60-69’,”>=70”))
table(dat$age, useNA = “always”)
#RIDRETH3- Race
dat$race[dat$RIDRETH3==’Mexican American’]<-“Hispanic”
dat$race[dat$RIDRETH3==’Other Hispanic’]<-“Hispanic”
dat$race[dat$RIDRETH3==’Non-Hispanic White’]<-“Non-Hispanic White”
dat$race[dat$RIDRETH3==’Non-Hispanic Black’]<-“Non-Hispanic Blacks”
dat$race[dat$RIDRETH3==’Non-Hispanic Asian’]<-“Non-Hispanic Asian”
dat$race[dat$RIDRETH3==’Other Race - Including Multi-Rac’]<-“Other Race”
dat$race[dat$RIDRETH3==’Missing’]<-NA
table(dat$race, useNA = “always”) summary(dat)
#DMDMARTL -Marital Status
dim(dat) names(dat)
dat$Marital [dat$DMDMARTL==’Widowed’]<-‘Previously Married’ dat$Marital [dat$DMDMARTL==’Divorced’]<-‘Previously Married’ dat$Marital [dat$DMDMARTL==’Separated’]<-‘Previously Married’ dat$Marital [dat$DMDMARTL==’Living with partner’]<-‘Married’ dat$Marital [dat$DMDMARTL==’Married’]<-‘Married’ dat$Marital [dat$DMDMARTL==’Never married’]<-‘Never Married’ dat$Marital [dat$DMDMARTL==’Refused’]<-NA dat$Marital [dat$DMDMARTL==’Missing’]<-NA dat$Marital <- na.omit(dat$Marital)
table(dat$Marital, useNA = “always”)
“INDFMPIR” #Ratio of family income to poverty #In general, a ratio less than 1 means that the income is less than the poverty level. When the ratio equals 1, the income and poverty level are the same, and when the ratio is greater than 1, the income is higher than the poverty level. #http://neocando.case.edu/cando/pdf/CensusPovertyandIncomeIndicators.pdf
dat$pir [dat$INDFMPIR <1]<-‘Below poverty level’ dat$pir [dat$INDFMPIR ==1 ]<-‘Poverty level’ dat$pir [dat$INDFMPIR >1]<-‘Above poverty level’ dat$pir [dat$INDFMPIR==’Missing’]<-NA
table(dat$pir, useNA = “always”)
“LBXSCA” #Total Calcium (mg/dL)
#REFERENCE RANGES (NORMAL VALUES) Calcium #Serum or Plasma Age Group #mg/dL
#>12 Y #8.5-10.5 #Reference Range values were established from wellness participants with an age mix similar to our patients. These data were analyzed using non-parametric techniques described by Reed (Clin Chem 1971;17:275) and Herrara (J Lab Clin Med 1958;52:34-42) which are summarized in recent editions of Tietz’ textbook. Descriptions appear in Clin Chem 1988;34:1447 and Clinics in Laboratory Medicine June 1993;13:481. #Pediatric Reference Range Guidelines for Synchron Systems- Multicenter study using data from Montreal, Quebec, Miami, FL and Denver, CO. Beckman 1995
dat$calc<-cut(dat$LBXSCA, c(-Inf,8.5,10.5, Inf),labels=c(‘low ca’,’normal ca’,”high ca”))
table(dat$calc, useNA = “always”)
“DSQTVD” #Vitamin D (D2 + D3) (mcg) #According to the Office of Dietary Supplements, the following are the minimum amounts of vitamin D a person needs per day: #https://ods.od.nih.gov/factsheets/VitaminD-Consumer/
#1–70 years 15 mcg to 600 IU #71+ years 20 mcg to 800 IU
dat$vitd<-cut(dat$DSQTVD, c(-Inf,15, Inf),labels=c(‘lowvitd’,”highvitd”))
table(dat$vitd, useNA = “always”)
#DIQ010 - Doctor told you have diabetes
dat$diab [dat$DIQ010==’Yes’]<-‘Yes’ dat$diab [dat$DIQ010==’No’]<-‘No’ dat$diab [dat$DIQ010==’Borderline’]<-‘No’ dat$diab [dat$DIQ010==”Don’t know”]<-NA
table(dat$diab, useNA = “always”)
#MCQ160A - Doctor ever said you had arthritis
dat$arth [dat$MCQ160A==’Yes’]<-‘Yes’ dat$arth [dat$MCQ160A==’No’]<-‘No’ dat$arth [dat$MCQ160A==”Don’t know”]<-NA dat$arth [dat$MCQ160A==’Missing’]<-NA
table(dat$arth, useNA = “always”)
#MCQ160f - Ever told you had a stroke
dat$strok [dat$MCQ160F==’Yes’]<-‘Yes’ dat$strok [dat$MCQ160F==’No’]<-‘No’ dat$strok [dat$MCQ160F==”Don’t know”]<-NA dat$strok [dat$MCQ160F==’Missing’]<-NA
table(dat$strok, useNA = “always”)
#WHD010 - Current self-reported height (inches)
#WHD020 - Current self-reported weight (pounds)
dat$bm <- (dat$WHD020) / (dat$WHD010 * dat$WHD010)*703
dat$bmi<-cut(dat$bm, c(-Inf,18.5,25, 30, Inf),labels=c(‘underweight’,’normalweight’,”overweight”, “obese”))
table(dat$bmi, useNA = “always”)
#ALQ130 - Avg # alcohol drinks/day - past 12 mos
dat$alc<-cut(dat$ALQ130, c(-Inf,1,20, Inf),labels=c(‘<1’,’1-19’,”>=20”)) table(dat$alc, useNA = “always”)
#ALQ111 - Ever had a drink of any kind of alcohol dat$alq [dat$ALQ111 ==’Yes’]<-‘Yes’ dat$alq [dat$ALQ111 ==’No’ ]<-‘No’ dat$alq [dat$ALQ111==’Missing’]<-NA
table(dat$alq, useNA = “always”)
#OSQ060 - Ever told had osteoporosis/brittle bones dat$osteo [dat$OSQ060 ==’Yes’]<-‘Yes’ dat$osteo [dat$OSQ060 ==’No’ ]<-‘No’ dat$osteo [dat$OSQ060==’Refused’]<-NA dat$osteo [dat$OSQ060==”Don’t know”]<-NA dat$osteo [dat$OSQ060==’Missing’]<-NA
table(dat$osteo, useNA = “always”)
#OSQ130 - Ever taken prednisone or cortisone daily dat$pred [dat$OSQ130 ==’Yes’]<-‘Yes’ dat$pred [dat$OSQ130 ==’No’ ]<-‘No’ dat$pred [dat$OSQ130==’Refused’]<-NA dat$pred [dat$OSQ130==”Don’t know”]<-NA dat$pred [dat$OSQ130==’Missing’]<-NA table(dat$pred, useNA = “always”)
#OSQ150 - Parents ever told had osteoporosis? dat$fmhx [dat$OSQ150 ==’Yes’]<-‘Yes’ dat$fmhx [dat$OSQ150 ==’No’ ]<-‘No’ dat$fmhx [dat$OSQ150==’Refused’]<-NA dat$fmhx [dat$OSQ150==”Don’t know”]<-NA dat$fmhx [dat$OSQ150==’Missing’]<-NA table(dat$fmhx, useNA = “always”)
#SMQ020 - Smoked at least 100 cigarettes in life dat$smk [dat$SMQ020 ==’Yes’]<-‘Yes’ dat$smk [dat$SMQ020 ==’No’ ]<-‘No’ dat$smk [dat$SMQ020==’Refused’]<-NA dat$smk [dat$SMQ020==”Don’t know”]<-NA dat$smk [dat$SMQ020==’Missing’]<-NA table(dat$smk, useNA = “always”)
#SMQ040 - Do you now smoke cigarettes?
dat$smkfreq [dat$SMQ040 ==’Every day’]<-‘Every day’ dat$smkfreq [dat$SMQ040 ==’Some days’ ]<-‘Some days’ dat$smkfreq [dat$SMQ040==’Not at all’]<- ‘Not at all’ dat$smkfreq [dat$SMQ040==”Don’t know”]<-NA dat$smkfreq [dat$SMQ040==’Missing’]<-NA table(dat$smkfreq, useNA = “always”)
#”DMDEDUC2”- Education level
dat$edu [dat$DMDEDUC2 ==’Less than 9th grade’]<-‘noHS’ dat$edu [dat$DMDEDUC2 ==”High school graduate/GED or equi”]<-‘some/completedHS’ dat$edu [dat$DMDEDUC2==”9-11th grade (Includes 12th grad”]<- ‘some/completedHS’ dat$edu [dat$DMDEDUC2==’Some college or AA degree’]<- ‘highereducation’ dat$edu [dat$DMDEDUC2==’College graduate or above’]<- ‘highereducation’ dat$edu [dat$DMDEDUC2==”Refused”]<-NA dat$edu [dat$DMDEDUC2==”Don’t know”]<-NA dat$edu [dat$DMDEDUC2==’.’]<-NA table(dat$edu, useNA = “always”)
#t score-(bmd-mean reference bmd)/sd It is the bone mineral density (BMD) at the site when compared to the young normal reference mean
dat$Tscorefemurneck <- (dat$DXXNKBMD-0.947) / 0.139 dat$Tscorespine <- (dat$DXXOSBMD -1.074) / 0.126
dat$Tscorefemurneck<-cut(dat$Tscorefemurneck, c(-Inf,-2.5,-1.0, Inf),labels=c(‘osteo’,’low bone mass’,”normal”)) table(dat$Tscorefemurneck, useNA = “always”)
dat$femurneckBMD<-dat$DXXNKBMD dat$SpineBMD<-dat$DXXOSBMD
#RHQ540 - Ever use female hormones? dat$horm [dat$RHQ540 ==’Yes’]<-‘Yes’ dat$horm [dat$RHQ540 ==’No’ ]<-‘No’ dat$horm [dat$RHQ540==’Refused’]<- NA dat$horm [dat$RHQ540==”Don’t know”]<-NA dat$horm [dat$RHQ540==’Missing’]<-NA table(dat$horm, useNA = “always”)
dat$vig [dat$PAQ605==’Yes’]<-‘Vigorous’ dat$vig [dat$PAQ605==”No”]<-‘NoVigorous’ dat$vig [dat$PAQ605==”Don’t Know”]<-NA dat$mod [dat$PAQ620==’Yes’]<-‘Moderate’ dat$mod [dat$PAQ620==”No”]<-‘NoModerate’ dat$mod [dat$PAQ620==”Don’t Know”]<-NA dat$vig [dat$PAQ650==’Yes’]<-‘Vigorous’ dat$vig [dat$PAQ650==”No”]<-‘NoVigorous’ dat$vig [dat$PAQ650==”Don’t Know”]<-NA dat$mod [dat$PAQ665==’Yes’]<-‘Moderate’ dat$mod [dat$PAQ665==”No”]<-‘NoModerate’ dat$mod [dat$PAQ665==”Don’t Know”]<-NA table(dat$vig) table(dat$mod)
dat$RIAGENDR<-NULL dat$RIDAGEYR<-NULL dat$RIDRETH3<-NULL dat$DMDMARTL<-NULL dat$INDFMPIR<-NULL dat$LBXSCA<-NULL dat$DSQTVD<-NULL dat$DIQ010<-NULL dat$MCQ160A<-NULL dat$MCQ160F<-NULL dat$WHD020<-NULL dat$WHD010<-NULL dat$OSQ010A<-NULL dat$OSQ010B<-NULL dat$OSQ010C<-NULL dat$ALQ130<-NULL dat$ALQ111<-NULL dat$OSQ060<-NULL dat$OSQ130<-NULL dat$SMQ020<-NULL dat$SMQ040<-NULL dat$OSQ010C<-NULL dat$ALQ130<-NULL dat$ALQ111<-NULL dat$OSQ060<-NULL dat$OSQ130<-NULL dat$OSQ150<-NULL
dat$DMDEDUC2<-NULL dat$bm<-NULL dat$RHQ540<-NULL dat$DXXNKBMD<-NULL dat$DXXOSBMD<-NULL dat$PAQ605<-NULL dat$PAQ620<-NULL dat$PAQ635<-NULL dat$PAQ650<-NULL dat$PAQ665<-NULL dat$PAD680<-NULL dat$Hipfrac<-NULL dat$Wristfrac<-NULL dat$spinefrac<-NULL dat$Spinefrac<-NULL
names(dat) ##########################################33333######################################################################################### #saving data for late use getwd() save(dat, file=”Project.RData”)
#loading saved data load(‘/Users/winnie/Desktop/project/finalProject.RData’) names(dat) summary(dat) dim(dat) glimpse(dat) str(dat)
DATA ENGINEERING
table(dat$race, useNA = “always”) summary(dat$frac) #View(dat$frac) #Observation 2166 frac value is NA dat<-dat[-2166,] lapply(dat[c(2:22,26:28)], unique) dat[,c(2:22,26:28)]<-lapply(dat[,c(2:22,26:28)],as.factor) dat[,c(23:25)]<-lapply(dat[,c(23:25)],as.numeric) table(dat$frac, useNA = “always”) lapply(dat[c(2:22,26:28)], unique) summary(dat) fit<-glm(frac~.-SEQN,dat,family=binomial) x<-dat dat[,c(“SEQN”,”vitd”,”alc”,”smkfreq”,”Tscorespine”,”SpineBMD”,”horm”, “pir”)]<-NULL #removing variables with missing values of more than 40% : vitd- 52%,”alc”- 42%,”smkfreq”- 54%,”Tscorespine”-49%,”SpineBMD”-49%,”horm- 55%, Pir-13% names(dat) dim(dat)
#missing values sum(is.na(dat)) 423 (18.5%)
mean(is.na(dat)) 0.009256018
pMiss <- function(x){sum(is.na(x))/length(x)*100} apply(dat,2,pMiss) apply(dat,1,pMiss)
#the remainig 20 variables has missing values less than 5% dat<-dat[-1273,] #removing observation missing more than 2 features #observation 1273 is missing 3 features dim(dat) [1] 2284 20
library(VIM)
aggr_plot <- aggr(dat, col=c(‘navyblue’,’red’), numbers=TRUE, sortVars=TRUE, labels=names(dat), cex.axis=.7, gap=1, ylab=c(“Histogram of missing data”,”Pattern”))
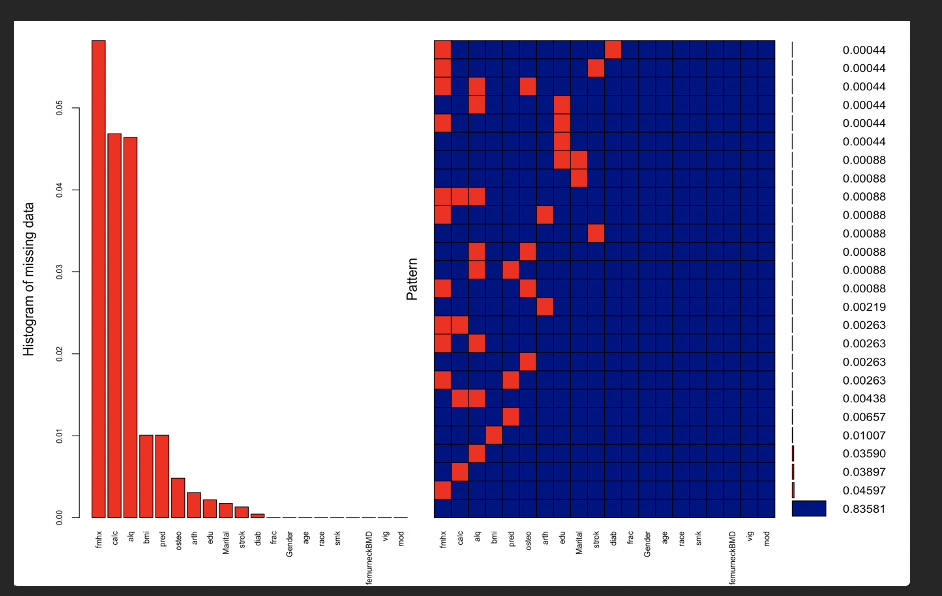
#In checking the patern of the missing values, 84% of the samples are not missing any information
write.csv(dat,”/Users/winnie/Desktop/ILE/Project.csv”, row.names = TRUE) #convert Rdata to CSV file)
require(mice) #initialize an empty model to take the parameters from empty_model <- mice(dat, maxit=0) method <- empty_model$method predictorMatrix <- empty_model$predictorMatrix
#first make a bunch of guesses… imputed_data <- mice(dat, method, predictorMatrix, m=5) #then pick one for each variable imputed_data <- complete(imputed_data)
head(imputed_data)
as.data.frame(imputed_data) # convert to data-frame
df <- imputed_data sum(is.na(df)) head(df) names(df) str(df)
Exploratory Data Analysis
summary (df$Gender) prop.table(table(df$Gender,df$frac)) table(df$arth,df$frac)
summary (df$femurneckBMD)
require(ggplot2) theme_few(base_size = 12, base_family = “”) ggplot(df,aes(age,fill=frac))+geom_bar(stat=”count”,position=”dodge”)
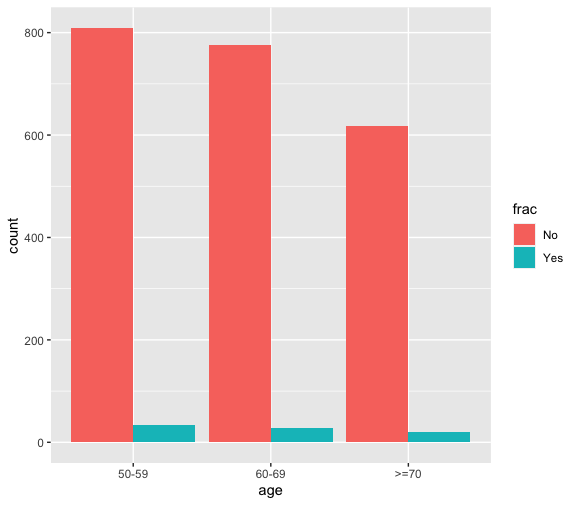
#there are more males thn fremale with fractures
Modelling
#Get a training and a testing set to use when building some models require(caret) set.seed(100) data_index <- createDataPartition(df$frac, p=0.7, list = FALSE) traindat <- df[data_index,] testdat <- df[-data_index,]
Applying machine learning models
#Set cross-validation and build and compare different predictive models fitControl <- trainControl(method=”cv”, number = 5, classProbs = TRUE, summaryFunction = twoClassSummary)
Logistics regression
fit<-glm(frac~.,data=traindat ,family=binomial(logit)) summary(fit)
fit1<-glm(frac~Gender+ calc+ arth+ alq+ osteo, data=traindat ,family=binomial(logit)) summary(fit1)
#Confusion Matrix and predictions pred_glm <- predict(fit, testdat, type=”response”) table(testdat$frac, pred_glm >0.5)
#accuracy print(“Accuracy”) (617+0)/(617+67+1+0) #0.9007
pred_glm1 <- predict(fit1, testdat, type=”response”) table(testdat$frac, pred_glm1 >0.5)
(618+0)/(618+67+0+0) #0.9022
#specificity print(“Specificity”) (0/(0+1)) #0
##recall and sensitivity print(“Recall and Sensitivity”) 617/(617+67) #0.90205
#precision print(“Precision”) 617/(617+1) #0.99838
#miss-classification print(“Miss-classification”) (67+1)/(67+617+1+0) #0.09927
Decision tree
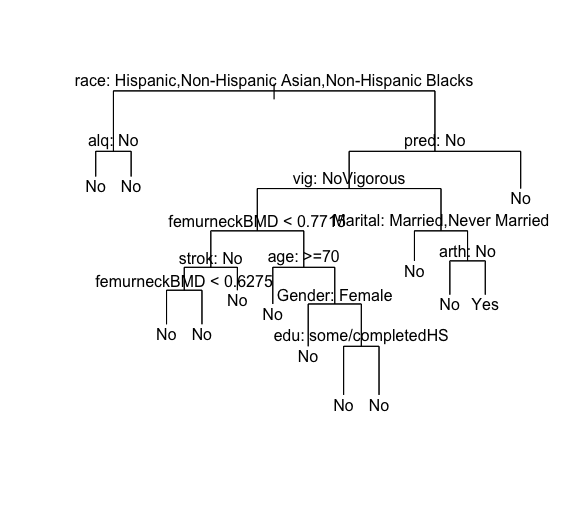
require(‘tree’) fit_cart = tree(frac~.,data=traindat, control=tree.control(nobs=nrow(traindat), mincut = 5)) summary(fit_cart)
#The decision tree has 3 terminal nodes and misclassified 0.098 individuals.
plot(fit_cart) text(fit_cart, pretty = 0)
#prune the tree using cross validation fit_cv = cv.tree(fit_cart,K=10, FUN = prune.misclass)
fit_cv plot(fit_cv)
fit_cv$dev ##[1] 1026.208 1038.904 1042.146 fit_cv$size ##[1] 3 2 1 min.ind = which.min(fit_cart_cv$dev) min.size = fit_cart_cv$size[min.ind] fit_cart_prune = prune.tree(fit_cart, best=min.size) plot(fit_cart_prune) text(fit_cart_prune,pretty = 0)
#The final classification tree model shows that race and femoral bmd is related to the fracture outcome
#prediction tree.pred = predict(fit_cart, testdat, type=”class”) with(testdat, table(tree.pred, testdat$frac)) (618+0)/687 Accuracy= 0.8995633
Random Forest
require(“randomForest”) model_rf <- train(frac~., data=traindat, method=”ranger”, metric=”ROC”, preProcess = c(‘center’, ‘scale’), na.action = na.pass, importance=TRUE, trControl=fitControl)
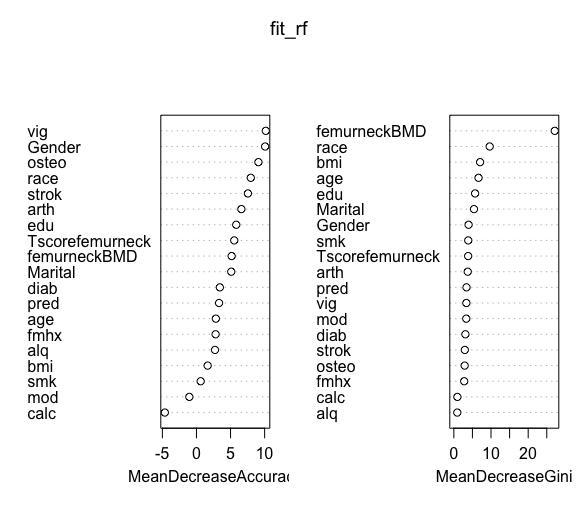
fit_rf = randomForest(frac~.,data=traindat, importance =TRUE, mtry=5, ntree=500)
varImpPlot(fit_rf)
pred_rf <- predict(fit_rf, testdat) cm_rf <- confusionMatrix(pred_rf, testdat$frac) cm_rf
#Get importance importance <- importance(fit_rf) varImportance <- data.frame(Variables = row.names(importance), Importance = round(importance[ ,’MeanDecreaseGini’],2))
#important variables- femoral neck bmd, Tscore femoral neck, calcium, gender, age, arthritis, BMI and smoking
Lasso/ Ridge Regression
require(glmnet) x = data.matrix(traindat[,2:20]) y = traindat$frac
set.seed(356) #10 fold cross validation cvfit.m.ridge = cv.glmnet(x, y, family = “binomial”, alpha = 0, type.measure = “class”)
cvfit.m.lasso = cv.glmnet(x, y, family = “binomial”, alpha = 1, type.measure = “class”)
plot(cvfit.m.lasso) par(mfrow=c(1,2)) plot(cvfit.m.ridge, main = “Ridge”) plot(cvfit.m.lasso, main = “Lasso”)
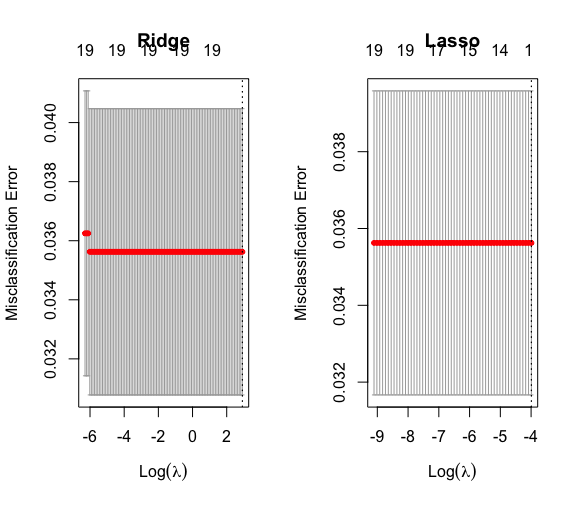
#Ridge gave a better missclassification error
coef(cvfit.m.ridge, s = “lambda.min”) round(coef(cvfit.m.ridge,s=”lambda.min”)[,1],2) round(coef(cvfit.m.lasso,s=”lambda.1se”))
yhat = rep(NA, nrow(x))
#prediction Pred.M = predict(cvfit.m.ridge, newx=data.matrix(testdat[,2:20]), type=”class”) table(testdat$frac, Pred.M)
#prediction Pred.l = predict(cvfit.m.lasso, newx=data.matrix(testdat[,2:20]), type=”class”) table(testdat$frac, Pred.l)
Neural network
model_lda_nnet <- train(frac~., traindat, method=”nnet”, metric=”ROC”, preProcess=c(‘center’, ‘scale’), tuneLength=10, trace=FALSE, na.action = na.pass, trControl=fitControl)
pred_lda_nnet <- predict(model_lda_nnet, testdat) cm_lda_nnet <- confusionMatrix(pred_lda_nnet, testdat$frac) cm_lda_nnet
SVM with radial kernel
model_svm <- train(frac~., traindat, method=”svmRadial”, metric=”ROC”, preProcess=c(‘center’, ‘scale’), na.action = na.pass, trace=FALSE, trControl=fitControl)
pred_svm <- predict(model_svm, testdat) cm_svm <- confusionMatrix(pred_svm, testdat$frac) cm_svm
LASSO
#we have to create a vector for potential lambdas lambda_vector <-10^seq(-5,5,length=500) set.seed(1992)
Model1 <-train(frac~.,data =traindat, method=”glmnet”, metric=”ROC”, tuneGrid=expand.grid(alpha=1,lambda=lambda_vector), trControl=fitControl, preProcess=c(“center”,”scale”), na.action = na.omit) Model1 round(coef(Model1$finalModel,Model1$bestTune$lambda),3)
#Plot log(lambda) & AUC plot(log(Model1$results$lambda),Model1$results$AUC, xlab=”Log(lambda)”, ylab=”AUC”, xlim=c(-10,100)) log(Model1$bestTune$lambda)
varImp(Model1) ggplot(varImp(Model1))+ labs(title = “Model Variable importance Rank from LASSO”)
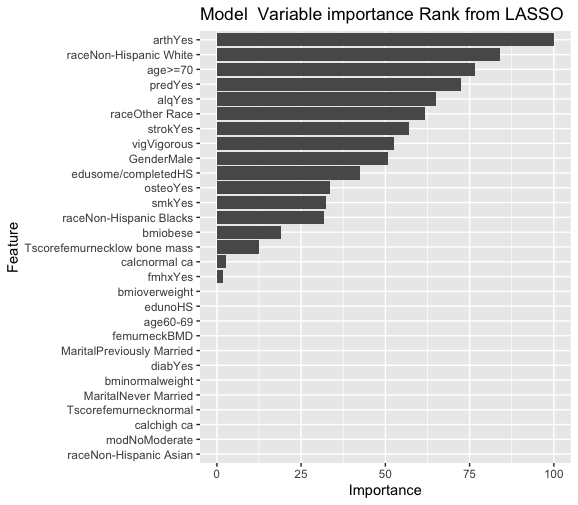
pred1 <- predict(Model1, testdat) cm1 <- confusionMatrix(pred1, testdat$frac) cm1
RANDOM FOREST with CV
rf_ctr_specs1 <-trainControl(method = “repeatedcv”, repeats = 2, number=5, classProbs = TRUE, search = “random”)
Model2.a <-train(frac~.,data=traindat, method=”rf”, metric=”ROC”, trControl=rf_ctr_specs1)
Model2.a dev.off() plot(varImp(Model2.a,scale=F),main=”RandomForest 5-FOLDS-2-REPEATED CV”) varImp(Model2.a)
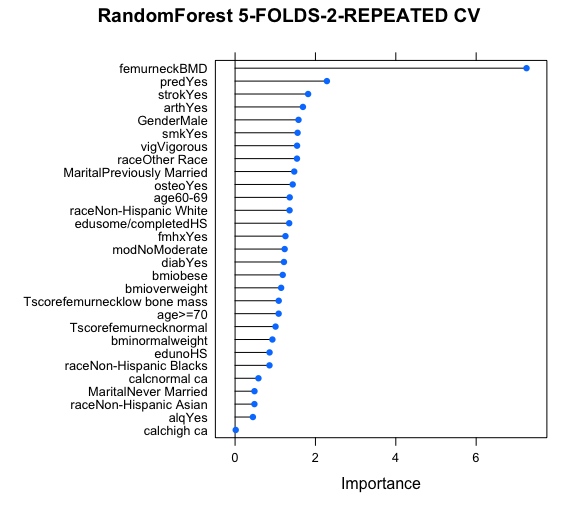
pred2 <- predict(Model2.a, testdat) cm2 <- confusionMatrix(pred2, testdat$frac) cm2
BAGGING
bag.df<-randomForest(frac~.,data=traindat,mtry=19,importance=T)
summary(bag.df) bag.df$importance varImpPlot(bag.df,col=rainbow(19))
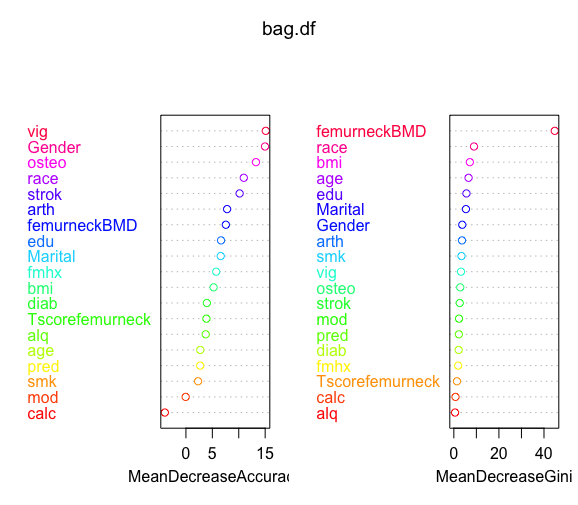
pred3 <- predict(bag.df, testdat) cm3 <- confusionMatrix(pred3, testdat$frac) cm3
BOOSTING
install.packages(“gbm”) library(gbm) set.seed(1992)
boost.df<-gbm(frac~.,data = traindat,distribution = “gaussian”, n.trees=5000,interaction.depth = 4) boost.df summary(boost.df)
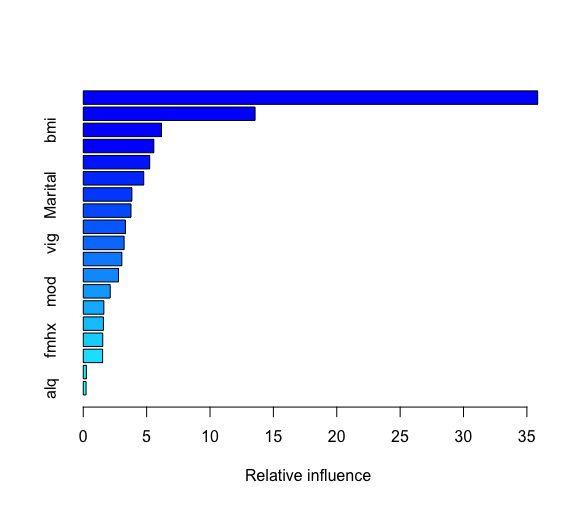
pred4 <- predict(boost.df, testdat, n.trees = 5000) cm4 <- confusionMatrix(pred3, testdat$frac) cm4
```
CONCLUSION
This study explores the use of machine learning methods in osteoporotic fracture prediction in the older population in US adults. The machine learning model can predict the osteoporotic fracture solely by demographic and clinical examination findings and this would help primary care physicians serving the rural and underserved communities to predict osteoporotic fractures of the hip, femur, and spine. If applied to a bigger data set, I believe the predictive accuracy of our model will further increase. We propose that machine learning is an important modality of the medical research field.