PubMed Abstract Retrieval COVID VACCINE
AIM- To extract covid vaccine abstracts from PubMed
Skills- Topic modeling, NLP, LDA, numpy, urlib, entrez, scikit_learn, HTTPError, wordcloud, pyLDAvis Gensim, coherence score, model perplexity, Interactive topic model visualization.
Introduction
Topic modeling is a form of unsupervised learning that identifies hidden relationships in data.Topic modeling is an area of natural language processing that can analyze text without the need for annotation—this makes it versatile and effective for analysis at scale. I used the Latent Dirichlet Allocation (LDA) approach in this topic modelling. (Giri, 2021)
A multinomial distribution is a generalization of the more familiar binomial distribution (which has 2 possible outcomes, such as in tossing a coin). A K-nomial distribution has K possible outcomes (such as in a K-sided dice).
In LDA, the Dirichlet is a probability distribution over the K-nomial distributions of topic mixes.By using a generative process and Dirichlet distributions, LDA can better genaralize to new documents after it’s been trained on a given set of documents.(Giri,2021)
import numpy
import urllib
import Bio
from Bio import Entrez
import time
from urllib.error import HTTPError # for Python 3
#!pip3 install biopython
Entrez.email = "oshaba3@uic.edu"
handle = Entrez.esearch(db="pubmed", term="COVID vaccine", reldate=365, datetype="pdat", usehistory="y")
result = Entrez.read(handle)
count = int(result["Count"])
print("Found %i results" % count)
result
batch_size = 10
out_handle = open("COVIDvaccine.txt", "w")
for start in range(0,count,batch_size):
end = min(count, start+batch_size)
print("Going to download record %i to %i" % (start+1, end))
attempt = 1
while attempt <= 3:
try:
fetch_handle = Entrez.efetch(db="pubmed",rettype="medline",
retmode="text",retstart=start,
retmax=batch_size,
webenv=result["WebEnv"],
query_key=result["QueryKey"])
attempt=4
except HTTPError as err:
if 500 <= err.code <= 599:
print("Received error from server %s" % err)
print("Attempt %i of 3" % attempt)
attempt += 1
time.sleep(15)
else:
raise
data = fetch_handle.read()
fetch_handle.close()
out_handle.write(data)
out_handle.close()
OPEN THE FILE
def extract_abstracts(in_file,out_file):
again=False
buffer=[]
for line in in_file:
if line.startswith("AB") or again==True and not (line.startswith('FAU') or line.startswith('CI')):
buffer.append(line)
again=True
continue
elif (line.startswith("FAU") or line.startswith("CI")) and again==True:
out_file.write("".join(buffer))
buffer=[]
again=False
else:
continue
file_in = open("COVIDvaccine.txt")
file_out = open("abstract_COVIDvaccine.txt","w")
extract_abstracts(file_in,file_out) ## extract the abstracts from the file
data=open('abstract_COVIDvaccine.txt').read().split('AB')
data
len(data)
data[1]
newdata=data[1:400]
newdata[1]
TEXT PREPROCESSING
- Tokenization, which breaks up text into useful units for analysis
- Normalization, which transforms words into their base form using lemmatization techniques (eg. the lemma for the word “studies” is “study”)
- Part-of-speech tagging, which identifies the function of words in sentences (eg. adjective, noun, adverb)
LOWER CASING/ GENERATING WORD CLOUD
#!pip3 install wordcloud
import pandas as pd
from wordcloud import WordCloud, STOPWORDS
import matplotlib.pyplot as plt
comment_words = ' '
stopwords = set(STOPWORDS)
# iterate through the the text file ( newdata)
for val in range(len(newdata)):
# typecaste each val to string
val = str(newdata[val])
# split the value
tokens = val.split()
# Converts each token into lowercase
for i in range(len(tokens)):
tokens[i] = tokens[i].lower()
comment_words += " ".join(tokens)+" "
wordcloud = WordCloud(width = 800, height = 800,
background_color ='white',
stopwords = stopwords,
min_font_size = 10).generate(comment_words)
# plot the WordCloud image
plt.figure(figsize = (8, 8), facecolor = None)
plt.imshow(wordcloud)
plt.axis("off")
plt.tight_layout(pad = 0)
plt.show()
TOKENIZATION/ LEMMATIZATION/ REMOVE STOP WORDS
# import all libraries
from nltk.corpus import stopwords
from nltk.stem.wordnet import WordNetLemmatizer
from nltk.stem.porter import PorterStemmer
import string
from nltk.tokenize import RegexpTokenizer
# set the stop word list, punctuation list and lemma
stop = set(stopwords.words('english'))
#You can add custom words in your stop list
stop.update(('and','I','A','And','So','arnt','This','When','It','many','Many'))
exclude = set(string.punctuation)
# initialize lemmatizer
lemma = WordNetLemmatizer()
# initialize stemmer
porter_stemmer = PorterStemmer()
# initialize tokenizer
tokenizer = RegexpTokenizer(r'\w+')
texts = []
# loop through document list
for i in newdata:
# clean and tokenize document string
raw = i.lower()
tokens = tokenizer.tokenize(raw)
# remove stop words from tokens
stopped_tokens = [i for i in tokens if not i in stop]
# stem tokens
stemmed_tokens = [porter_stemmer.stem(i) for i in stopped_tokens]
# add tokens to list
texts.append(stemmed_tokens)
texts[0]
DATA TRANSFORMATION/ CORPUS AND DICTIONARY
#!pip3 install gensim
# Using Gensim library turn our tokenized documents into a id <-> term dictionary
# For all the documents, a corpus always contains each word’s token’s id along with its frequency count in the document.
import gensim
from gensim import corpora, models
dictionary = corpora.Dictionary(texts)
dictionary.save('dictionary.dict')
print(dictionary)
dictionary[1]
# convert tokenized documents into a document-term matrix ( Bag of words model)
BoW_corpus = [dictionary.doc2bow(text) for text in texts]
corpora.MmCorpus.serialize('corpus.mm', BoW_corpus)
print (len(BoW_corpus))
#
print (BoW_corpus[3])
The above output shows that the word with id=0 appears 3 times in the document 0. The above output is not readable by human. We can also convert these ids to words but for this we need our dictionary to do the conversion as follows In [29]:
id_words = [[(dictionary[id], count) for id, count in line] for line in BoW_corpus]
print(id_words[3])
MODELING
import pandas as pd
pd.DataFrame(id_words )
# Initialize the model
tfidf = models.TfidfModel(BoW_corpus)
corpus_tfidf = tfidf[BoW_corpus]
for doc in corpus_tfidf:
print(doc)
pd.DataFrame(corpus_tfidf)
tf_idf_id_words = [[(dictionary[id], count) for id, count in line] for line in corpus_tfidf]
tf_idf_id_words[0]
lda_model = gensim.models.ldamodel.LdaModel(
corpus=corpus_tfidf, id2word=dictionary, num_topics=20, random_state=100,
update_every=1, chunksize=100, passes=10, alpha='auto', per_word_topics=True
)
lda_model
print(lda_model.print_topics())
doc_lda = lda_model[corpus_tfidf]
COMPUTING MODEL PERPLEXITY and COHERENCE SCORE
print('\nPerplexity: ', lda_model.log_perplexity(corpus_tfidf))
The Model Perplexity is how good the model is. The lower the score the better the model will be
from gensim.models import CoherenceModel
# Compute Coherence Score
coherence_model_lda = CoherenceModel(model=lda_model, texts=texts, dictionary= dictionary, coherence='c_v')
coherence_lda = coherence_model_lda.get_coherence()
print('\nCoherence Score: ', coherence_lda)
INTERACTIVE TOPIC MODEL VISUALIZATION
#!pip3 install pyLDAvis
import pyLDAvis
import pyLDAvis.gensim
import matplotlib.pyplot as plt
pyLDAvis.enable_notebook()
vis = pyLDAvis.gensim.prepare(lda_model, corpus_tfidf, dictionary)
vis
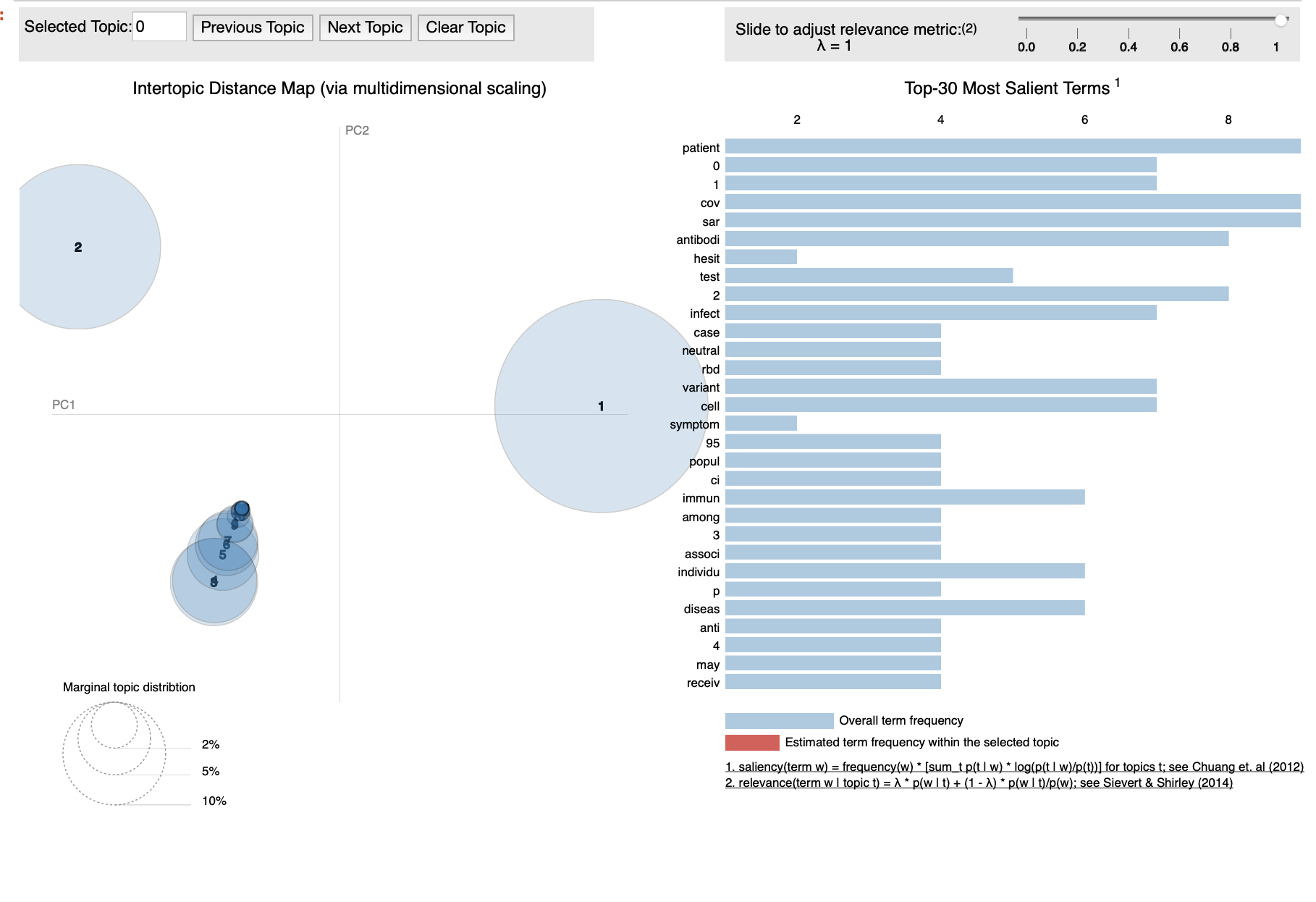
From the above output, the bubbles on the left-side represents a topic and larger the bubble, the more prevalent is that topic. The topic model is good because the topic model has big, non-overlapping bubbles scattered throughout the chart.